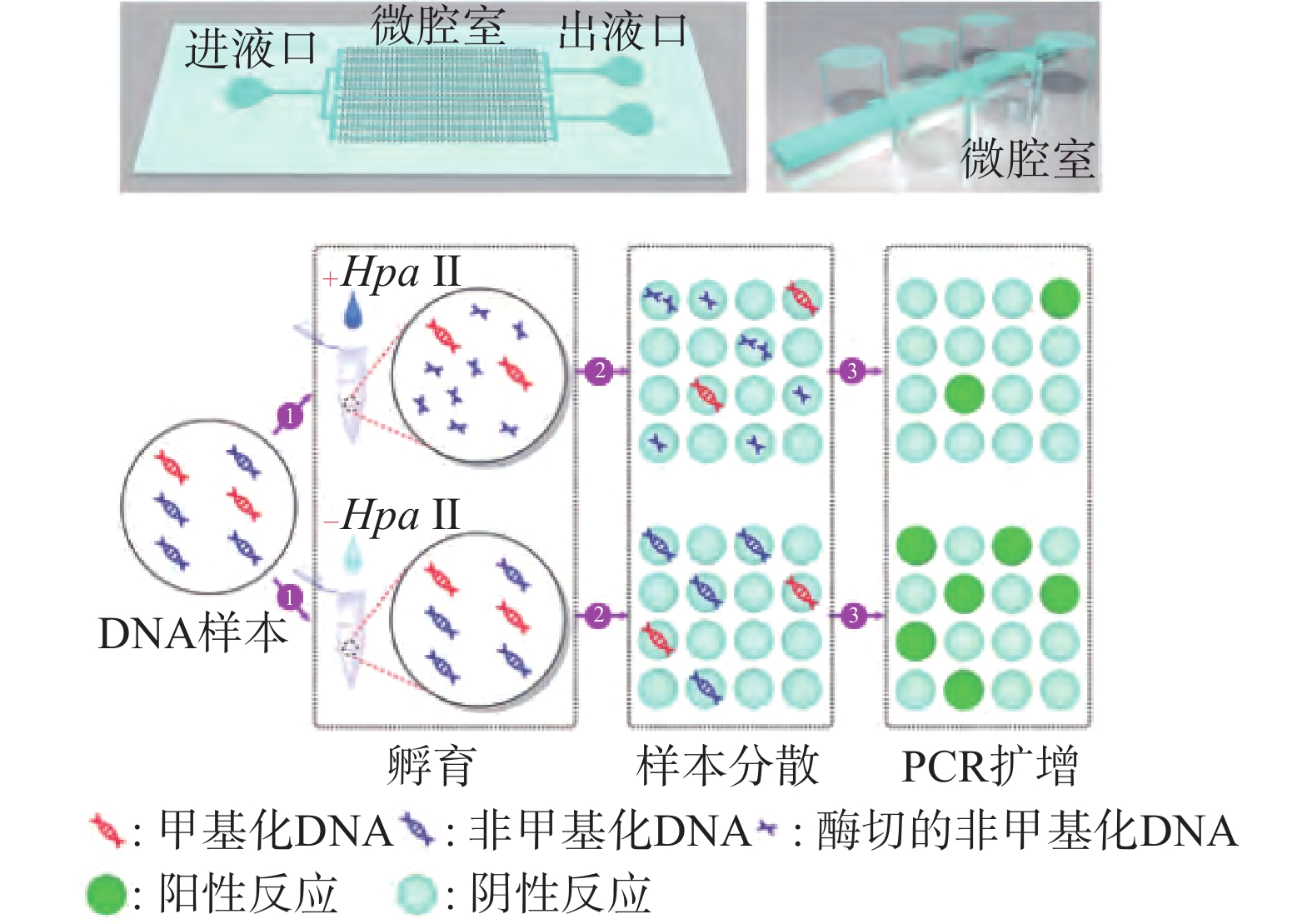
| [1] |
Jin Z, Liu Y. DNA methylation in human diseases[J]. Genes & diseases, 2018, 5(1): 1-8.
|
| [2] |
Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics[J]. Science, 2001, 293(5532): 1068-1070. doi: 10.1126/science.1063852
|
| [3] |
Shen L, Song CX, He C, et al. Mechanism and function of oxidative reversal of DNA and RNA methylation[J]. Annu Rev Biochem, 2014, 83: 585-614. doi: 10.1146/annurev-biochem-060713-035513
|
| [4] |
Schubeler D. Function and information content of DNA methylation[J]. Nature, 2015, 517(7534): 321-326. doi: 10.1038/nature14192
|
| [5] |
Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes[J]. Journal of molecular biology, 1987, 196(2): 261-282. doi: 10.1016/0022-2836(87)90689-9
|
| [6] |
Cross SH, Charlton JA, Nan X, et al. Purification of CpG islands using a methylated DNA binding column[J]. Nature genetics, 1994, 6(3): 236-244. doi: 10.1038/ng0394-236
|
| [7] |
Robertson K D. DNA methylation and human disease[J]. Nature reviews Genetics, 2005, 6(8): 597-610. doi: 10.1038/nrg1655
|
| [8] |
Kulis M, Esteller M. DNA methylation and cancer[J]. Advances in genetics, 2010, 70: 27-56.
|
| [9] |
How KA, Nielsen HM, Tost J. DNA methylation based biomarkers: practical considerations and applications[J]. Biochimie, 2012, 94(11): 2314-2337. doi: 10.1016/j.biochi.2012.07.014
|
| [10] |
Van DVL, Speeckaert R, Van G D, et al. DNA methylation-based biomarkers in serum of patients with breast cancer[J]. Mutation research, 2012, 751(2): 304-325. doi: 10.1016/j.mrrev.2012.06.001
|
| [11] |
Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges[J]. Nature reviews Genetics, 2012, 13(10): 679-692. doi: 10.1038/nrg3270
|
| [12] |
Šestáková Š, Šálek C, Remešová H. DNA Methylation Validation Methods: a Coherent Review with Practical Comparison[J]. Biological procedures online, 2019, 21: 19-25. doi: 10.1186/s12575-019-0107-z
|
| [13] |
王玲, 彭付端, 赵慧, 等. 焦磷酸测序和MassARRAY定量检测DNA甲基化在年龄推断中的差异[J]. 南方医科大学学报, 2020, 40(12): 145-151.
|
| [14] |
Feng L, Lou J. DNA Methylation Analysis[J]. Methods Mol Biol, 2019, 1894: 181-227.
|
| [15] |
Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method[J]. Biology, 2016, 5(3): 1-22.
|
| [16] |
Nakagawa T, Wakui M, Hayashida T, et al. Intensive optimization and evaluation of global DNA methylation quantification using LC-MS/MS[J]. Analytical and bioanalytical chemistry, 2019, 411(27): 7221-7231. doi: 10.1007/s00216-019-02115-3
|
| [17] |
杨佳怡, 傅博强, 李曼莉, 等. 基于DNA提取效率校正的数字PCR定量方法[J]. 计量科学与技术, 2020(10): 32-36. doi: 10.3969/j.issn.2096-9015.2020.10.09
|
| [18] |
Torres AL, Barrientos EY, Wrobel K, et al. Selective derivatization of cytosine and methylcytosine moieties with 2-bromoacetophenone for submicrogram DNA methylation analysis by reversed phase HPLC with spectrofluorimetric detection[J]. Analytical chemistry, 2011, 83(20): 7999-8005. doi: 10.1021/ac2020799
|
| [19] |
Gowher H, Leismann O, Jeltsch A. DNA of Drosophila melanogaster contains 5-methylcytosine. The EMBO journal[J]. 2000, 19(24): 6918-6923.
|
| [20] |
曹雷, 郭利娟, 郭晓锦, 等. 癌症液体活检新思路: 数字PCR检测DNA甲基化[J]. 生物化学与生物物理进展, 2019, 46(11): 50-65.
|
| [21] |
Laird PW. Principles and challenges of genomewide DNA methylation analysis[J]. Nature reviews Genetics, 2010, 11(3): 191-203. doi: 10.1038/nrg2732
|
| [22] |
Waalwijk C, Flavell RA. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations[J]. Nucleic acids research, 1978, 5(12): 4631-4634. doi: 10.1093/nar/5.12.4631
|
| [23] |
Kaput J, Sneider TW. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions[J]. Nucleic acids research, 1979, 7(8): 2303-2322. doi: 10.1093/nar/7.8.2303
|
| [24] |
Gautier F, Bünemann H, Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences[J]. European journal of biochemistry, 1977, 80(1): 175-183. doi: 10.1111/j.1432-1033.1977.tb11869.x
|
| [25] |
Vander P LH, Flavell RA. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroiqd tissues[J]. Cell, 1980, 19(4): 947-958. doi: 10.1016/0092-8674(80)90086-0
|
| [26] |
Hayatsu H, Wataya Y, Kazushige K. The addition of sodium bisulfite to uracil and to cytosine[J]. J Am Chem Soc, 1970, 92(3): 724-726. doi: 10.1021/ja00706a062
|
| [27] |
Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis-a personal account[J]. Proceedings of the Japan Academy Series B, 2008, 84(8): 321-330. doi: 10.2183/pjab.84.321
|
| [28] |
Herman JG, Graff JR, Myöhänen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(18): 9821-9826. doi: 10.1073/pnas.93.18.9821
|
| [29] |
Scher MB, Elbaum MB, Mogilevkin Y, et al. Detecting DNA methylation of the BCL2, CDKN2A and NID2 genes in urine using a nested methylation specific polymerase chain reaction assay to predict bladder cancer[J]. The Journal of urology, 2012, 188(6): 2101-2107. doi: 10.1016/j.juro.2012.08.015
|
| [30] |
Husseiny MI, Kuroda A, Kaye AN, et al. Development of a quantitative methylation-specific polymerase chain reaction method for monitoring beta cell death in type 1 diabetes[J]. PLoS One, 2012, 7(10): 47942. doi: 10.1371/journal.pone.0047942
|
| [31] |
Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate[J]. Science, 1998, 281(5375): 363-365.
|
| [32] |
Busato F, Dejeux E, El Abdalaoui H, et al. Quantitative DNA Methylation Analysis at Single-Nucleotide Resolution by Pyrosequencing[J]. Methods Mol Biol, 2018, 1708: 427-445.
|
| [33] |
Kunze S. Quantitative Region-Specific DNA Methylation Analysis by the EpiTYPER™ Technology[J]. Methods Mol Biol, 2018, 1708: 515-535.
|
| [34] |
Suchiman HE, Slieker RC, Kremer D, et al. Design, measurement and processing of region-specific DNA methylation assays: the mass spectrometry-based method EpiTYPER[J]. Frontiers in genetics, 2015, 6: 287-294.
|
| [35] |
Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry[J]. Proc Natl Acad Sci, 2005, 102(44): 15785-15790. doi: 10.1073/pnas.0507816102
|
| [36] |
Coolen MW, Statham AL, Gardiner-Garden M, et al. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements[J]. Nucleic acids research, 2007, 35(18): 119-125. doi: 10.1093/nar/gkm662
|
| [37] |
Ehrich M, Turner J, Gibbs P, et al. Cytosine methylation profiling of cancer cell lines[J]. Proc Natl Acad Sci, 2008, 105(12): 4844-4849. doi: 10.1073/pnas.0712251105
|
| [38] |
Docherty SJ, Davis OS, Haworth CM, et al. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation[J]. Epigenetics & chromatin, 2009, 2(1): 3-10.
|
| [39] |
Hussmann D, Hansen LL. Methylation-Sensitive High Resolution Melting (MS-HRM)[J]. Methods Mol Biol, 2018, 1708: 551-571.
|
| [40] |
Frommer M, McDonald LE, Millar, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands[J]. Proc Natl Acad Sci, 1992, 89(5): 1827-1831. doi: 10.1073/pnas.89.5.1827
|
| [41] |
Hsu HK, Weng YI, Hsu PY, et al. Detection of DNA methylation by MeDIP and MBDCap assays: an overview of techniques[J]. Methods Mol Biol, 2014, 1105: 61-70.
|
| [42] |
Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome[J]. Nature genetics, 2007, 39(4): 457-466. doi: 10.1038/ng1990
|
| [43] |
Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells[J]. Nature genetics, 2005, 37(8): 853-862. doi: 10.1038/ng1598
|
| [44] |
Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis[J]. Nature biotechnology, 2008, 26(7): 779-785. doi: 10.1038/nbt1414
|
| [45] |
Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells[J]. Nature genetics, 2006, 38(2): 149-153. doi: 10.1038/ng1719
|
| [46] |
Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis[J]. Cell, 2006, 126(6): 1189-1201. doi: 10.1016/j.cell.2006.08.003
|
| [47] |
Zilberman D, Gehring M, Tran RK, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription[J]. Nature genetics, 2007, 39(1): 61-69. doi: 10.1038/ng1929
|
| [48] |
Zhao MT, Whyte JJ, Hopkins GM, et al. Methylated DNA immunoprecipitation and high-throughput sequencing (MeDIP-seq) using low amounts of genomic DNA[J]. Cellular reprogramming, 2014, 16(3): 175-184. doi: 10.1089/cell.2014.0002
|
| [49] |
Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome[J]. Nucleic acids research, 2010, 38(2): 391-399. doi: 10.1093/nar/gkp992
|
| [50] |
Robinson MD, Stirzaker C, Statham AL, et al. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation[J]. Genome research, 2010, 20(12): 1719-1729. doi: 10.1101/gr.110601.110
|
| [51] |
Weisenberger DJ, Trinh BN, Campan M, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight[J]. Nucleic acids research, 2008, 36(14): 4689-4698. doi: 10.1093/nar/gkn455
|
| [52] |
Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples[J]. Nature biotechnology, 2009, 27(9): 858-863. doi: 10.1038/nbt.1559
|
| [53] |
Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers[J]. Proc Natl Acad Sci, 2017, 114(28): 7414-7419. doi: 10.1073/pnas.1703577114
|
| [54] |
Van W L, Janssens L, Meeuws H, et al. Droplet digital PCR is an accurate method to assess methylation status on FFPE samples[J]. Epigenetics, 2018, 13(3): 207-213. doi: 10.1080/15592294.2018.1448679
|
| [55] |
Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer[J]. Annals of oncology:official journal of the European Society for Medical Oncology, 2015, 26(9): 1994-1999. doi: 10.1093/annonc/mdv272
|
| [56] |
Cui X, Cao L, Huang Y, et al. In vitro diagnosis of DNA methylation biomarkers with digital PCR in breast tumors[J]. The Analyst, 2018, 143(13): 3011-3020. doi: 10.1039/C8AN00205C
|
| [57] |
Wu Z, Bai Y, Cheng Z, et al. Absolute quantification of DNA methylation using microfluidic chip-based digital PCR[J]. Biosensors & bioelectronics, 2017, 96: 339-344.
|
| [58] |
Ladd AC, Aryee MJ, Ordway JM, et al. Comprehensive high-throughput arrays for relative methylation (CHARM)[J]. Current protocols in human genetics, 2010, 20: 1-19.
|
| [59] |
Warnecke PM, Stirzaker C, Melki JR, et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA[J]. Nucleic acids research, 1997, 25(21): 4422-4426. doi: 10.1093/nar/25.21.4422
|
| [60] |
Warnecke PM, Stirzaker C, Song J, et al. Identification and resolution of artifacts in bisulfite sequencing[J]. Methods, 2002, 27(2): 101-107. doi: 10.1016/S1046-2023(02)00060-9
|
| [61] |
Hernández HG, Tse MY, Pang SC, et al. Optimizing methodologies for PCR-based DNA methylation analysis[J]. Biotechniques, 2013, 55(4): 181-197. doi: 10.2144/000114087
|
| [62] |
Sant KE, Nahar MS, Dolinoy DC. DNA methylation screening and analysis[J]. Methods Mol Biol, 2012, 889: 385-406.
|
| [63] |
Wang RY, Gehrke CW, Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues[J]. Nucleic acids research, 1980, 8(20): 4777-4790. doi: 10.1093/nar/8.20.4777
|
| [64] |
Pajares MJ, Palanca BC, Urtasun R, et al. Methods for analysis of specific DNA methylation status[J]. Methods, 2021, 187: 3-12. doi: 10.1016/j.ymeth.2020.06.021
|
| [65] |
陈桂芳, 欧阳艳艳, 杨佳怡, 等. 核酸标准物质测量方法研究进展[J]. 计量科学与技术, 2021, 65(6): 25-33. doi: 10.12338/j.issn.2096-9015.2020.9022
|
| [66] |
Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells[J]. Nature, 2008, 454(7205): 766-770. doi: 10.1038/nature07107
|
| [67] |
Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences[J]. Nature, 2009, 462(7271): 315-322. doi: 10.1038/nature08514
|
| [68] |
高运华, 陈鸿飞, 盛灵慧, 等. DNA甲基化定量测量国际比对CCQM P94.2[J]. 中国测试, 2015(6): 47-51. doi: 10.11857/j.issn.1674-5124.2015.06.011
|
| [69] |
牛春艳, 张永卓, 杨佳怡, 等. 基于数字PCR的质粒核酸标准物质合作定值研究[J]. 计量学报, 2021, 42(11): 1522-1527. doi: 10.3969/j.issn.1000-1158.2021.11.18
|
| [70] |
Yang I, Kim SK, Burke DG, et al. An international comparability study on quantification of total methyl cytosine content[J]. Analytical biochemistry, 2009, 384(2): 288-295. doi: 10.1016/j.ab.2008.09.036
|

 作者投稿
作者投稿 专家审稿
专家审稿 编辑办公
编辑办公


 下载:
下载: